In a recent publication on the bioRxiv* preprint server, researchers from ETH Zurich, Imperial College London, and Karolinska Institutet learned of increased resistance to antibody-mediated neutralization through severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) sublineage Omicron BAArray2. 75. 2.
The emerging sublines of SARS-CoV-2 Omicron host mutations in genes encoding various pico protein residues, resulting in increased immune evasion. Omicron BA. 4. 6 is lately a generalized lineage and carries the R346T and N658S mutations, while the emerging subline BA. 2. 10. 4 harbors mutations at position 486.
Several recent studies have shown further evasion of antibody-mediated neutralization through Omicron variants, adding lines BA. 5 and BA. 1. Specifically, mutations in the peak 346 residue have been linked to increased immune evasion.
Given the rapidly emerging variants with mutations that facilitate antibody leakage, it is vital to frequently check the effectiveness of recently used monoclonal antibodies compared to those variants to prevent recurrence of severe symptoms of coronavirus disease 2019 (COVID-19).
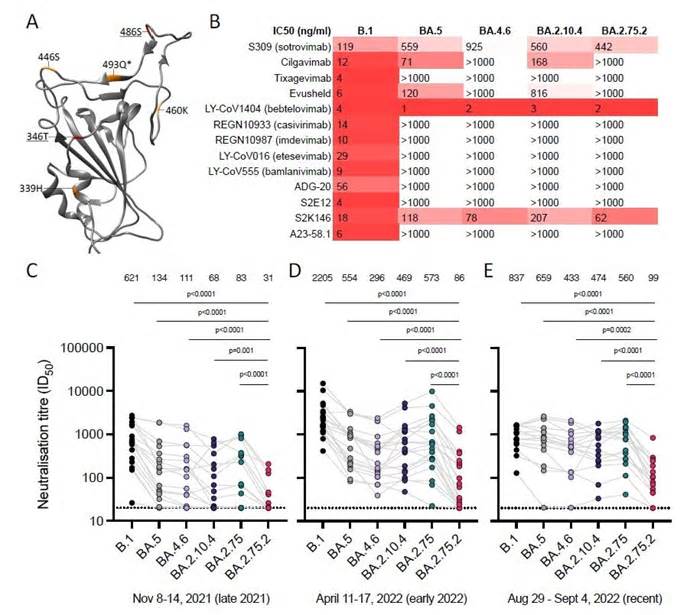
The provider tested the neutralization sensitivity of 3 sublines BA. 4. 6, BA. 2. 10. 4 and BA. 2. 75. 2 of Omicron, as opposed to a set of preclinical and clinically used monoclonal antibodies and recently donated blood serum. The researchers used targeted mutagenesis at multiple sites of ba. 4, ba. 2 and BA. 2. 75 expression plasmids to produce pseudoviruses for the sublines BA. 4. 6, BA. 2. 10. 4 and BA. 2. 75. 2.
Human embryonic kidney cells 293 (HEK293T) expressing human angiotensin-converting enzyme 2 (ACE2) were used to check neutralization sensitivity. In addition, pseudovirus neutralization controls were performed with a panel of monoclonal antibodies, adding bebtélovimab, cilgavimab and tixagevimab. Neutralization power of serum antibodies was achieved heat-inactivated serums. In addition, geometric mean neutralization titers (GMT) were also calculated to compare the neutralization sensitivity of other sublines.
The effects imply that the sublines BA. 4. 6 and BA. 2. 75. 2 completely escaped neutralization through cilgavimab and the Evusheld vaccine, a mixture of the two monoclonal antibodies cilgavimab and tixagevimab. The BA. 2. 10. 4 subline showed reduced sensitivity to cilgavimab. However, bebtélovimab able to neutralize all sublines. Sotrovimab showed a low neutralization efficacy compared to the 3 sublines tested in the study, as well as against the BA. 5 subline.
When tested against recently donated blood serum, BA. 2. 10. 4 and BA. 4. 6 (311 and 356 GMT, respectively) showed increased resistance to neutralization to the globally dominant BA. 5 subline (GMT of 453). Subline BA. 2. 75. 2 showed a GMT price five times lower than subline BA. 5, indicating a higher resistance to neutralization.
To conclude, the study tested the neutralizing strength of monoclonal antibodies in clinical use and preclinical trials against 3 emerging sublines of the Omicron variant of SARS-CoV-2, namely BA. 4. 6, BA. 2. 10. 4 and BA. 2. 75Array2. The efficacy of the serum in neutralizing these sublines was also tested.
The widely used monoclonal antibodies, cilgavimab and tixagevimab, which were used separately and in mixture as evusheld vaccine, showed virtually no efficacy against the 3 sublines. The only monoclonal antibody capable of neutralizing all sublines tested bebtélovimab. Serum antibodies also showed a reduction in neutralization force opposite to BA. 4. 6 and BA. 2. 10. 4. Subline BA. 2. 75. 2 showed the largest neutralization leak.
Overall, the effects imply that the emerging sublines of omicron bring mutations that decorate its ability to evade humoral immunity, highlighting the need for monoclonal antibodies and antiviral methods.
bioRxiv publishes initial clinical reports that are not peer-reviewed and therefore inconclusive, clinical consulting practices/health-related behaviors, nor are they treated as established information.
Written by
Chinta Sidharthan is a bangalore-founded India. Su academic background is in evolutionary biology and genetics, and has extensive experience in clinical studies, teaching, clinical writing and herpetology. Chinta holds a PhD in Evolutionary Biology from the Indian Institute of Science and is passionate about science education, writing, animals, wildlife and conservation. For his doctoral studies, he explored the origins and diversification of blind snakes in India, where he carried out extensive fieldwork in the jungles of southern India. He has won the Governor-General’s Bronze Medal and the Gold Medal for Academic Excellence from bangalore University and has published his studies in high-impact journals.
Use one of the following to cite this article in your essay, article, or report:
ap
Sidharthan, Chinta. (2022, September 21). Study of evasion of superior immunity through Omicron BA. 2. 75. 2. Actualités-Médical. Retrieved September 25, 2022, https://www. news-medical. net/news/20220921/Study–higher-immunity-evasion-through-Omicron-BA2752. aspx.
deputy
Sidharthan, Chinta. ” He shows further evasion of immunity through Omicron BA. 2. 75. 2″. News-Medical. September 25, 2022. .
Chicago
Sidharthan, Chinta. ” He shows further evasion of immunity through Omicron BA. 2. 75. 2″. News-Medical. https://www. news-medical. net/news/20220921/Study-shows-higher-immunity-evasion -via Omicron-BA2752. aspx. (accessed September 25, 2022).
Harvard
Sidharthan, Chinta. 2022. Superior immunity evasion study via Omicron BA. 2. 75. 2. News-Medical, accessed September 25, 2022, https://www. news-medical. net/news/20220921/Study–higher-immunity-evasion-by-Omicron-BA2752. aspx.
News-Medical. net – An AZoNetwork website
Ownership and operation through AZoNetwork, © 2000-2022