Researchers in the United States have made a vital discovery about the remedy and presentation of viral coronavirus 2 (SARS-CoV-2) peptides of severe acute respiratory syndrome that provides a greater understanding of the immune reaction to coronavirus disease 2019 (COVID-19) and would possibly help facilitate the progression of progressive vaccines.
The team presented the first detailed viral peptide research presented through class I human leukocyte antigen (HLA-I) in human cells inflamed with SARS-CoV-2.
Interestingly, researchers found that nine of the viral peptides are derived from non-canonical open reading frames (ORFs) in the viral protein Spike and nucleoprotein. Peak is the surface design that SARS-CoV-2 uses to bind to and infect host cells, while nucleoprotein is involved in the packaging of viral RNA and virions.
The team says the discovery suggests that T-cell reaction studies in patients with COVID-19 who lately only in canonical viral ORFs exclude a significant amount of HLA-I epitopes derived from the virus.
Current groups of HLA-I peptides derived from SARS-CoV-2 that identify viral signatures that activate T cells may be incomplete, say Pardis Sabeti (Broad Institute of MIT and Harvard, Cambridge) and his colleagues.
New approaches that directly identify naturally presented SARS-CoV-2 peptides would indicate the design of long-term sets to provide a more detailed characterization of T-cell responses in PATIENTS with COVID-19. Such approaches would also facilitate the design of more effective vaccines. the researchers say.
You must have a preprinted edition of the article on the bioRxiv server, while the article is peer-reviewed.
As study efforts continue to expand effective sarS-CoV-2 vaccines, it is to discern the role T cells play in building long-term immunity to the virus.
After infection, viral proteins are processed and found on the surface of the mobile host with HLA-I to be identified by cytotoxic T mobiles, which then induce an immune reaction to fight the virus.
To date, most studies that have tested the interaction between T cells and SARS-CoV-2 antigens have used bioinformatics predictions of HLA-I binding affinity. However, this technique is limited because it does not take into account all the steps involved in the remedy and presentation of the antigen, and the average positive predictive values received through the HLA alleles are still only about 64%.
These prediction models also don’t take into account how viruses can simply adjust cellular processes and antigen presentation.
”For example, viruses can reduce the translation of host proteins, proteasome machinery down and interfere with HLA-I expression’,” explain Sabeti and his team. the immune system. ‘
Experimental measurements of SARS-CoV-2 peptides presented in host cells are essential to understand how the immune formula responds to infection.
HLA-I immunodynamics based on mass spectrometry is a direct and impartial technique for finding endogenously presented peptides and building the processes that govern the remedy and presentation of the antigen.
“Taking advantage of HLA-I immunoopeptidoma datasets to be informed about virus-specific antigen processing regulations will improve our ability to, as it should be, expect viral epitopes and use them to examine immune responses in PATIENTS with COVID-19. “Array the researchers write.
Knowledge analysis derived from mass spectrometry requires the variety of a set of viral ORFs to examine, and so far those analyses have basically focused on canonical ORFs.
However, over the past decade, genome-wide sequencing studies have revealed an unexpected number of non-canonical ORFs in viral genomes, whose service remains largely unknown.
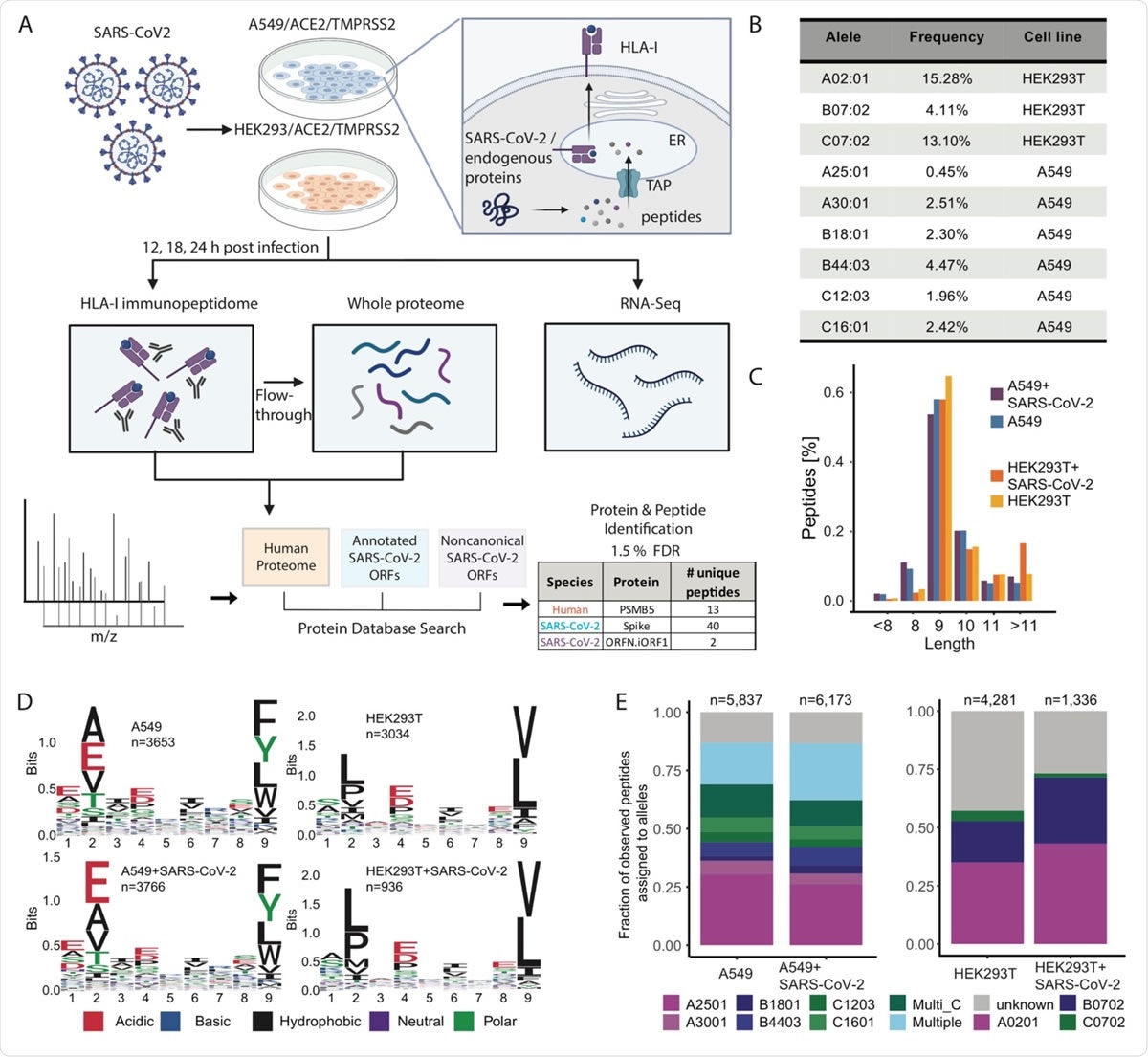
Sabeti and his colleagues have now presented the first detailed research of HLA-I immunoopeptidoma on two inflamed human moving lines with SARS-CoV-2 mass spectrometry, RNA sequencing and global proteomic measures.
The team met viral HLA-I peptides derived only from canonical ORF, but also from non-canonical ORF in SARS-CoV-2 Spike and nucleoprotein that are captured through existing vaccines.
“Surprisingly, nine of the two nine viral peptides detected are derived from out-of-frame internal ORFs in S [Spike] (S. iORF1/2) and N [nucleoprotein] (ORFnineb),” the researchers write.
The team says this discovery implies that existing studies examining T mobile responses in COVID-19 patients that only in canonical viral ORFs exclude a significant number of HLA-I epitopes derived from the virus.
Researchers estimated that on the list of endogenously treated and endogenously treated SARS-CoV-2 peptides that they identified, a group of 24 peptides would provide one or more peptides for at least one HLA-I allele in 99% of the human population.
Sabeti and his colleagues say the effects of the test can be used to facilitate a variety of data-driven peptides for immune surveillance and for the design of effective vaccines.
“These effects recommend that existing peptide sets may be incomplete,” the researchers write.
Consequently, unsewn approaches such as LC-MS/MS immunodynamics that directly identify SARS-CoV-2 peptides naturally presented on the host’s mobiles would possibly indicate the design of long-term sets to allow a more complete characterization of T-cell responses in COVID 19 patients,” the team concluded.
bioRxiv publishes initial clinical reports that are not peer reviewed and should therefore not be considered conclusive, clinical practices of consultation/health-related behaviors, nor treated as established information.
Written by
Sally holds a bachelor’s degree in biomedical sciences (B. Sc. ). He specializes in reviewing and synthesizing the latest discoveries in all medical spaces covered in high-impact foreign medical journals, foreign press meetings, and announcements from government agencies and regulators. At News-Medical, Sally generates news, life science articles, and interview coverage.
Use one of the following to cite this article in your essay, job, or report:
Apa
Robertson, Sally. (2020, 05 October). New findings can only examine the reaction of T cells in COVID-19. News-Medical. Recovered October 19, 2020 in https://www. news-medical. net/news/20201005/New-findings-may just– T-to-COVID-19. aspx-cell-reaction-reaction-research.
Mla
Robertson, Sally. ” New discoveries can simply look for the reaction of T cells in COVID-19. “News-Medical. 19 October 2020.
Chicago
Robertson, Sally. ” New discoveries can simply examine the reaction of T cells in COVID-19. “News-Medical. https: //www. news-medical. net/news/20201005/New-findings-may just– T-to-COVID-19. aspx-cell-reaction-research. (accessed 19 October 2020).
Harvard
Robertson, Sally. 2020. De new findings can examine the reaction of T cells in COVID-19. News-Medical, see October 19, 2020, https://www. news-medical. net/news/20201005/New -the-findings-can-only-research-of-cell-reaction-T-to-COVID-19. aspxArray
News-Medical. net – An AZoNetwork site
Ownership and operation through AZoNetwork, © 2000-2020